New simulations speedily account for a greater range of experimental parameters and show promise to resolve new levels of molecular detail.
From the Journal: The Journal of Chemical Physics
WASHINGTON, D.C., August 14, 2018 – Researchers from the University of Florida and the Federal University of Mato Grosso do Sul in Brazil have used state of the art simulations to assess the effect of both pH and redox potential, or rate of electron transfer, on a biomolecule. Additionally, Vinícius Cruzeiro, one of the authors on the paper, significantly reduced the required computational processing time using graphics processing unit (GPU) hardware.
“We’ve brought super computing power to the desktop, and have accelerated the simulation process from a couple of months to half a day for a new system,” said Adrian Roitberg, another author on the paper. This technology can now be used anywhere in the world for as little as about $500. The newly developed methodology is described in The Journal of Chemical Physics, from AIP Publishing.
In the mid-1970s, computers were programmed as “computational microscopes” to simulate experimental outcomes for biomolecules. The idea was that if information from computations could be sufficiently accurate, the simulations would provide a complete representation of the system in atomic detail. Over the years, researchers honed simulations to near experimental conditions, but before now the redox potential of biomolecules could not be accounted for. This metric describes a huge number of electrons transferred in cells throughout the human body.
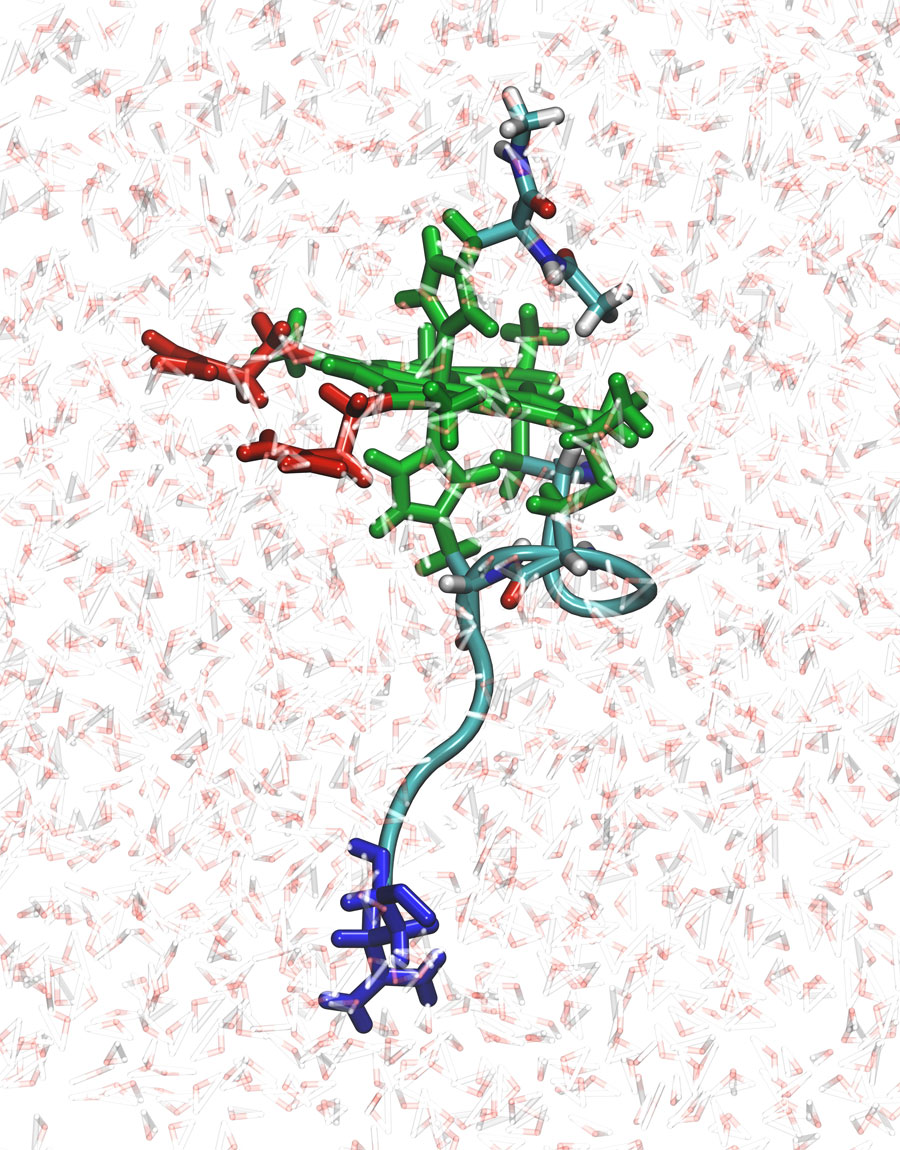
The group simulated the pH and redox potentials within a single peptide — N-acetylmicroperoxidase (NAcMP8) — of cytochrome c, a biomolecule in the respiratory chain containing essential redox centers called heme groups. “Heme proteins are present all over biology in redox processes; carrying oxygen from one place to another in the body, and in enzymes in the liver degrading toxins,” Roitberg said.
Experimentalists had already tested the effect of changes in pH and redox potential on NAcMP8, but Cruzeiro implemented new combined computational methods to simulate these results. Roitberg is a co-developer of the AMBER (Assisted Model Building with Energy Refinement) software suite, and it was within this software that Cruzeiro developed the new molecular dynamics methods.
It took hundreds of thousands of steps for Cruzeiro to simulate the outcome of changes in pH or redox potential for each individual residue in the peptide. On completion, Roitberg said he was “gladly surprised” to see that the simulations matched the experimental results.
Simulations like these traditionally require substantial time using super computers. However, Cruzeiro was able to create new replica exchange methods and implemented them on modern GPUs to speed up the process. In replica exchange programs, many simulations run simultaneously with crosstalk between them, effectively “team-working” to boost computational efficiency.
The ability to see the atomic level detail of redox reactions through the computational microscope will enable scientists to better understand a whole host of biomolecules. This opens the door in biomedicine to more directly detect and address disease at the molecular level.
“Now we’re looking at systems in the respiratory chain with four hemes, and finding out which heme gets the electron first, as the order is absolutely crucial. Experimentally it’s hard to say which of the four, but on the computational microscope we can figure it out,” Roitberg said.
###
For More Information:
Rhys Leahy
media@aip.org
301-209-3090
@AIPPhysicsNews
Article Title
Authors
Vinícius Cruzeiro, Marcos S. Amaral and Adrian E. Roitberg
Author Affiliations
University of Florida and Federal University of Mato Grosso do Sul, Campo Grande, Brazil
