Computational modeling of swimming shows the generation of fingerlike projections that give unilateral movement of amoeboid cells
From the Journal: Physics of Fluids
 WASHINGTON, D.C., October 31, 2017 — Rhythmic patterns and precise motions — these are key elements of proper swimming. Olympians demonstrate repeated patterns of breathing, with synchronized head, leg and arm movements, enthralling spectators and provoking applause for record-breaking paces. Comparable demonstrations of this pattern repetition and power usage can also be seen in a microscopic swimmer — the amoeboid cell.
WASHINGTON, D.C., October 31, 2017 — Rhythmic patterns and precise motions — these are key elements of proper swimming. Olympians demonstrate repeated patterns of breathing, with synchronized head, leg and arm movements, enthralling spectators and provoking applause for record-breaking paces. Comparable demonstrations of this pattern repetition and power usage can also be seen in a microscopic swimmer — the amoeboid cell.
The cell swimming shapes are now predictable to new levels of precision thanks to advanced 3-D modeling. Researchers Eric J. Campbell and Prosenjit Bagchi, from the Mechanical and Aerospace Engineering Department at Rutgers University, generated a 3-D model of an amoeba practicing pseudopod-driven swimming. The research will appear on the cover of this month’s issue of Physics of Fluids, from AIP Publishing.
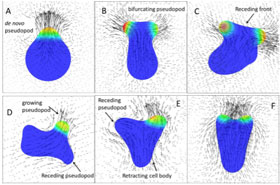
Amoeboid cells have uniquely flexible cytoskeletons, with no set shape. They can contract and expand their skeletal system while simultaneously altering the consistency of their cytoplasm, the plasma surrounding the cell’s organelles. Amoeboid cells are also distinctive with their capacity for pseudopod-driven motility. Pseudopods, meaning false feet, are projections of the cell body that can grow, split or retract to provide locomotion. Pseudopod movement is more complex than most would expect. It relies on biomolecular reactions, cell deformation and the motion of both cytoplasm and extracellular fluid.
“In this research, we combined a state-of-the-art model for cell deformation with intra- and extra-cellular fluid motion, and protein biochemistry using a dynamic pattern formation model,” Campbell said. “We then used parallel supercomputers to predict the cell’s motion, and studied its behavior by varying cell deformability, fluid viscosity, and protein diffusivity.”
Amoeboid cells show a unidirectionality in swimming with a corresponding change in pseudopod dynamics, caused by projections becoming more prevalent at the front of the cell. This unidirectionality is likely caused by an increased swimming speed due to the focused orientation. Using computer model simulations, the researchers studied cell swimming by varying protein diffusivity, membrane elasticity and cytoplasmic viscosity.
Accurately modeling amoeboid cell swimming posed a number of challenges. “The model needed to be able to resolve deformation in three-dimensions with high accuracy and without any numerical instability,” Campbell said. Protein biochemistry, which creates the locomotive force, had to be coupled to the model. Fluid motion also had to be considered. “The intra- and extra-cellular fluids may have dissimilar properties, and the model must account for such differences.”
These various parameters were integrated to optimize cell locomotion modeling, providing new, more precise information about the locomotion mechanisms. Amoeboid cells demonstrating this pseudopod-driven motility can offer insights into many biological processes. According to Campbell, the mobility is also observed during embryonic development, wound healing, immune response by white blood cells, and metastatic cancer cells.
###
For More Information:
Julia Majors
media@aip.org
301-209-3090
@AIPPhysicsNews
Article Title
A computational model of amoeboid cell swimming
Authors
Eric J. Campbell and Prosenjit Bagchi
Author Affiliations
Rutgers University
