A group of researchers is showing how artificial intelligence approaches can help identify cause-effect relationships within data.
From the Journal: The Journal of Chemical Physics
WASHINGTON, D.C., April 17, 2018 — Machine learning algorithms excel at finding complex patterns within big data, so researchers often use them to make predictions. Researchers are pushing this emerging technology beyond finding correlations to help uncover hidden cause-effect relationships and drive scientific discoveries.
At the University of South Florida, researchers are integrating machine learning techniques into their work studying proteins. As they report in The Journal of Chemical Physics, from AIP Publishing, one of their main challenges has been a lack of methods to identify cause-effect relationships in data obtained from molecular dynamics simulations.
“Proteins can be thought of as nanoscopic machines that perform a set of tasks. But when and where proteins carry out their specific tasks is controlled by cells through various stimuli, such as small molecules,” said Sameer Varma, an associate professor of biophysics at USF. “These stimuli interact with proteins to switch them ‘on’ and ‘off,’ and can even modify their speeds and strengths.”
In most proteins, the biological stimuli interact with a site on the protein that’s relatively far away from the part that carries out its corresponding task, requiring a signaling pathway. “This remote-control manner of switching in proteins is known as ‘allosteric signaling.’ Many proteins of pharmaceutical significance have now been identified where the dynamics or the ‘jiggling and wiggling’ of their constituent atoms are known to be vital to allosteric signaling,” Varma said. “The details, however, remain sketchy.”
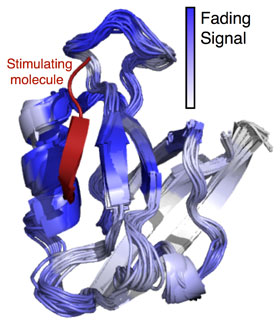 Varma and colleagues believe machine learning approaches can make a difference. “Developing and using machine learning techniques will enable us to find cause-effect relationships in protein dynamics data and begin to finally address some of the very fundamental questions in protein allostery,” he said. “One of our key findings was that the signal initiated at the stimulation site of the protein appeared to weaken as it moved away from the stimulation site. It came as a surprise, because no distance dependence was observed for the coupling of thermal motions between protein sites.”
Varma and colleagues believe machine learning approaches can make a difference. “Developing and using machine learning techniques will enable us to find cause-effect relationships in protein dynamics data and begin to finally address some of the very fundamental questions in protein allostery,” he said. “One of our key findings was that the signal initiated at the stimulation site of the protein appeared to weaken as it moved away from the stimulation site. It came as a surprise, because no distance dependence was observed for the coupling of thermal motions between protein sites.”
The group’s work demonstrates how machine learning approaches can be used to identify cause-effect relationships within data. Beyond this, “these techniques are allowing us to plug critical gaps in protein allostery,” Varma said. “Ultimately, when our methods are applied to the many proteins of pharmaceutical interest, we expect the mechanistic details to reveal much-needed new intervention strategies for restoring protein activities in diseased states. The general biophysical insights we gain should also help to inspire novel biomimetic solutions for many nanoengineering problems, such as nanosensor design for targeted drug delivery.”
The researchers envision exciting new work that will grow from their recent findings. “So far, we’ve focused on equilibrium data, but the signaling process has a critical nonequilibrium component that we haven’t explored yet,” Varma said. The group also plans to explore the role of the surrounding waters in signaling in greater detail, as well as apply their machine learning techniques to a wide set of protein families to determine the extent to which their new biophysical findings are generalizable.
###
For More Information:
Julia Majors
media@aip.org
301-209-3090
@AIPPhysicsNews
Article Title
Authors
Mohsen Botlani, Ahnaf Siddiqui and Sameer Varma
Author Affiliations
University of South Florida
