The AMOEBA force field can distinguish selective ion binding in calcium and magnesium ions by incorporating quantum methods.
From the Journal: The Journal of Chemical Physics
WASHINGTON, D.C., October 17, 2017 — Muscle twitching, lactose digestion, blood movement — what could possibly connect these body functions? You may be surprised to learn that all these processes and many more are driven by metal ions.
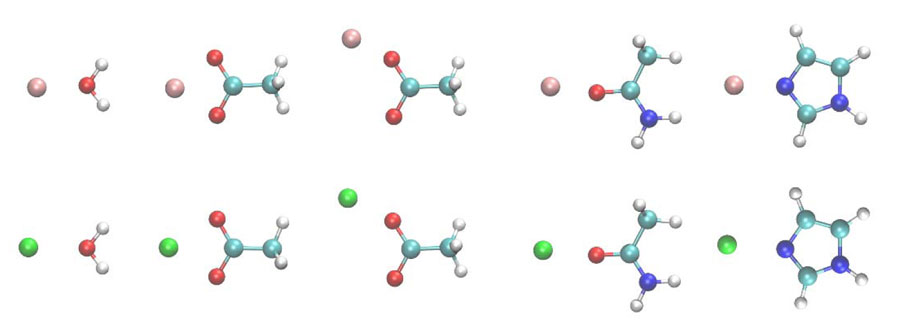
Sodium (Na+), potassium (K+), calcium (Ca2+) and magnesium (Mg2+) might be household names, but they are also critical to the function of human cells. The combination of these ions with bodily proteins creates complexes that are imperative to our continued existence.
The importance of proteins and metal ion interactions is well understood, but the mechanistic interactions between the two are still far from a complete picture.
Zhifeng Jine, Rui Qi, Chengwen Liu and Pengyu Ren, professors in the biomedical engineering department at the University of Texas at Austin, are working to quantitatively describe protein-ion interactions using what is called an atomic multipole optimized energetics for biomolecular applications (AMOEBA) force field. They describe their work in this week’s The Journal of Chemical Physics, from AIP Publishing.
AMOEBA technology was developed by Ren and Jay Ponder, from the University of Texas at Austin and Washington University in St. Louis. It uses a polarizable atomic multipole model to calculate the potential energy of a system. The AMOEBA model aims to address known inadequacies of current force field technology to accurately model ion and protein interactions.
“The lack of detailed understanding is mainly due to the lack of accurate and yet computationally efficient models to treat metal ions,” Ren said. “We aim to apply high-level quantum mechanical methods and advanced force field simulations to understand the nature of the interactions between metal ions and proteins.”
AMOEBA improves on classically based biomolecular simulations, whose models represent the intermolecular interaction using the sum of two forces, according to Ren: the van der Waals interaction and the Coulomb interaction between point charges.
“This representation is simple enough to be computationally tractable, and it has been widely used over the last few decades,” he said. “However, metal ions can lead to strong polarization and charge transfer effects, which are missing in these classical models. We believe these effects play [a] critical role in the specific ion-protein interaction.”
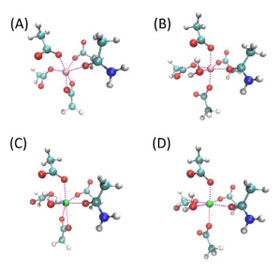
Ren and his colleagues looked specifically at the interaction of Mg2+/Ca2+ and amino acids. Magnesium and calcium are some of the most abundant ions in metalloproteins, proteins with a metal ion cofactor. Both are selectively bound to specific amino acids, making them interesting target molecules. The many-body response, the mutual induction between ions and their surrounding residues within the protein binding pockets, also impacts the binding affinity of these ion and can be captured by the AMOEBA model.
“The difference between the many-body response to Ca2+ vs Mg2+ in the binding pocket is substantial,” said Ren. “It was known that polarization and charge-transfer are important in protein-ion complexes, but for the competitive binding, many have suspected that these effects might cancel out.”
The AMOEBA model and the advancements of force fields, including applications of these results, Ren pointed out, are relevant in many diseases including cancer and neurodegenerative disorders. Knowledge of protein-ion interactions can provide foundational understanding for the advancement in related medical researches.
 ###
###
For More Information:
Julia Majors
media@aip.org
301-209-3090
@AIPPhysicsNews
Article Title
Authors
Zhifeng Jing, Rui Qi, Chengwen Liu and Pengyu Ren
Author Affiliations
University of Texas at Austin
