Researchers have combined existing methods to optimize simulations and better reflect the physical process of protein folding.
From the Journal: The Journal of Chemical Physics
 WASHINGTON, D.C., September 4, 2018 — A protein’s folding patterns help them perform their dedicated tasks. As the real “doers” of the cell, even a tiny alteration in a protein’s amino acid backbone can cause misfolding and hinder the protein’s functionality or cause disease. For instance, if tau, a protein that helps stabilize the structure of brain cells, is misfolded, it can form tau-tangles, which are commonly seen in Alzheimer’s patients.
WASHINGTON, D.C., September 4, 2018 — A protein’s folding patterns help them perform their dedicated tasks. As the real “doers” of the cell, even a tiny alteration in a protein’s amino acid backbone can cause misfolding and hinder the protein’s functionality or cause disease. For instance, if tau, a protein that helps stabilize the structure of brain cells, is misfolded, it can form tau-tangles, which are commonly seen in Alzheimer’s patients.
Scientists seek to better understand protein folding to cure misfolding diseases, but this incredibly complex process requires sophisticated algorithms to identify the folding mechanisms. Computational biophysicists from the Tata Institute of Fundamental Research Hyderabad (TIFR-H) have proposed a new way to identify the most crucial factors for protein folding. They demonstrated the short simulation time of their approach on a small but intriguing protein, “GB1 beta-hairpin,” in The Journal of Chemical Physics, from AIP Publishing.
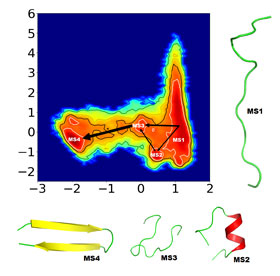
“By combining a method known as ‘Time-structure based independent component analysis’ (TICA) with short molecular dynamics simulations, we’ve found four physically meaningful intermediate folding states, not previously observed, and showed helical states which cannot usually be detected by other methods,” said Navjeet Ahalawat, an author on the paper.
Each atom in a protein can fold in three dimensions, but with millions of atoms present in even simple proteins, the task of understanding the collective folding combination becomes convoluted. Scientists have considered the different factors influencing protein folding, such as hydrogen bonding, and combined these into general descriptions called collective variables (CVs). However, with lots of potential factors, scientists lack a good way to find CVs that appropriately describe a feasible process.
“There are many ways proteins can go from unfolded to folded states, so the most challenging thing is deciding where to start,” Ahalawat said. Jagannath Mondal, another author on the paper, added that it was easy to “get lost in the data.”
The team decided to study the externally protruding hairpin of the GB1 protein because of the large body of existing work and many potential folding possibilities already estimated in past CVs. Ahalawat and Mondal took a number of existing GB1 CVs as constituent CVs and linearly combined them using TICA to identify a pair of “optimized” CVs. Then, they input the optimized CVs into the Markov State Model and identified four intermediate folding states along with the possible connecting pathways.
“We asked, what are the features estimated previously for this particular protein that might really play a key role in the system? And can we find the right combination of conditions?” Ahalawat said. “In our work we can now quantitatively tell if that feature is at all relevant to the process.”
“Using short simulations, we have come up with the weight that you really need to use in a combination, and this gives the right folding pattern for a protein,” Mondal added. “It’s a really cheap way of figuring out protein folding.”
In their method, data from previous studies are needed to identify optimal CVs. The team envisions their technique can be used to uncover the internal mechanism of healthy protein folding to correct disease causing misfolded proteins. They also want to further develop their CV optimization method and apply them in biomolecular recognition and drug discovery. “In the future we plan to incorporate nonlinear methods, using neural-network based deep learning techniques to improve our model,” Ahalawat said.
###
For More Information:
Rhys Leahy
media@aip.org
301-209-3090
@AIPPhysicsNews
Article Title
Authors
Navjeet Ahalawat and Jagannath Mondal
Author Affiliations
Tata Institute of Fundamental Research, Hyderabad, India
