A team of researchers in Italy developed a cost-effective system to detect in-flow biomolecules in real-time using ultrasensitive fluorescence detection of microRNAs with spectrally encoded microgels
From the Journal: Biomicrofluidics
WASHINGTON, D.C., December 19, 2016 — Single-stranded, noncoding micro-ribonucleic acids (microRNAs), consisting of 18-23 nucleotides, play a key role in regulating gene expression. Levels of microRNAs circulating within blood can be correlated to different states of diseases such as cancer, neurodegenerative disorders and cardiovascular conditions. Many microRNAs within the blood are encapsulated within exosomes, nanoscale vesicles released by the cells.
Accurate measurement of the quantity of microRNAs circulating within the blood is extremely challenging because of their short lengths, similar sequences and low concentration levels. Due to their small number of nucleotides, traditional polymerase chain reaction (PCR) detection methods must necessarily involve a ligation, or linking, step to produce longer complementary DNA strands. Such ligation often produces large biases.
Consequently, large volumes of clinical samples typically required to obtain accurate measurements, but few conventional detection systems can handle this directly without proper sample preparation and volume reduction.
A team of researchers in Italy from the Istituto Italiano di Tecnologia and the University of Naples Federico II, both in Naples, set out to develop a simple, ultrasensitive fluorescence detection system of in-flow microRNAs that uses spectrally encoded microgels.
As the team reports in Biomicrofluidics, from AIP Publishing, until now such a multiplexed barcode detection approach has only been performed in time-consuming observation procedures, significantly hindering its possible diagnostic performance.
“Our technological achievement rests upon the straightforward implementation of a seemingly real-time, microfluidic-based readout of microRNA sequences of interest, handling down to a few microliters of target volume,” explained Filippo Causa, an associate professor of industrial bioengineering in the Department of Chemical, Materials Engineering, and Industrial Production at the University of Naples Federico II. “No previous RNA sequence amplification is required, which reduces evident sources of measurement errors.”
To do this, the researchers first explored a cost-effective and biocompatible non-Newtonian fluid to create the optimal 3-D alignment of microgels in the center of a square-shaped glass capillary.
They then used a simple microfluidic layout to flow the microgel and allow a continuous measurement of the fluorescence signal with several emission wavelengths for the multiplexed barcode detection.
“We chose microgels with non-overlapping fluorescence-emitting molecules designed to distinguish spectral barcodes for multiplex analysis … and to obtain an absolute quantification of microRNA sequences,” said Causa. “The precise microgel alignment at various throughput rates and an automatic microRNA sequence intensity normalization in flow gives us an opportunity to obtain reliable measurements, similar to quiescent measurement results, without any fundamental pretreatments of the measurement sample.”
To prove their concept of this multiplex spectral microgel analysis within a microfluidic flow, the team used “different barcodes corresponding to different emissions at specific wavelengths and the fluorescence intensity of known microRNA concentration,” which was measured for calibrations of the specific microRNA being explored. Causa said, “So far, nine different microgel barcodes have been tested in flow with our detection approach, and more codes are being prepared to multiplex it further.”
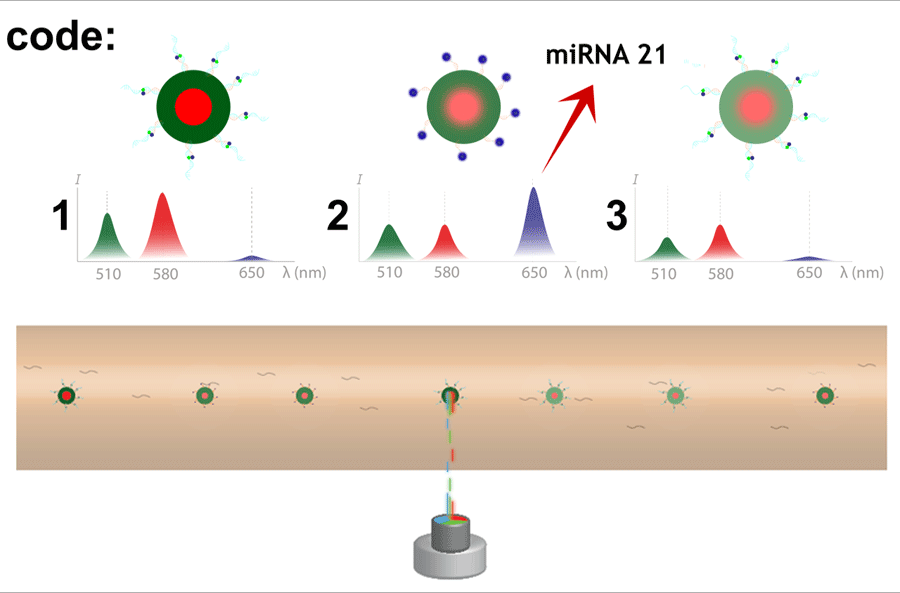
As a proof of principle, the team explored microRNA based on its significance to the pathogenesis of various malignant tumors including prostate, gastric, colon, breast and lung cancers.
“We were able to specifically detect, count and identify in a quasi-real-time manner hundreds of microgels (~80 microgel particles per minute) at sample volumes of only a few microliters,” said Causa. “Our system achieved a microRNA detection limit of 202 femtoMolars in microfluidic flow conditions.”
Measurements were performed with different microgel barcodes and one in particular focused on specific microRNA targets, demonstrating the specificity of the assay for multiplex measurement conditions.
“A microRNA 21 concentration of 0.74 picoMolars was detected in flow, which is consistent with the initial sample concentration level,” Causa said. “Out of such fluorescence acquisitions, an absolute quantification of the microRNA 21 concentration level was possible.”
In terms of applications for the system, since the specific target detection of microgels can be easily tuned, it can be applied to a wide range of different biomarkers thanks to its barcode structure.
“Users can also easily adjust its readout speed specifically for any microscopic system,” said Causa. “This means that the system will open up new options for biosensing particles within microfluidic devices.”
###
For More Information:
AIP Media Line
media@aip.org
301-209-3090
Article Title
In-flow real-time detection of spectrally encoded microgels for miRNA absolute quantification
Authors
David Dannhauser, Filippo Causa, Edmondo Battista, Angela M. Cusano, Domenico Rossi and Paolo A. Netti
Author Affiliations
Istituto Italiano di Tecnologia and the University of Naples Federico II
