Scientists have created a novel two-layer microchip to track individual developmental changes in stem cells over longer time periods.
From the Journal: Biomicrofluidics
WASHINGTON, D.C., September 17, 2019 — An interdisciplinary team has found a solution to a problem plaguing developmental biology — long-term cell tracking and manipulation.
Mechanical engineer Adela Ben-Yakar, at the University of Texas at Austin, collaborated with stem cell biologist Joshua Brickman, from the University of Copenhagen in Denmark, to painstakingly develop an automated microfluidic device for the stable imaging of mice embryonic stem cells over a three-day period. They describe the device and its utility in Biomicrofluidics, from AIP Publishing.
“Looking at single cells and seeing how, depending on their initial state, they respond, enables us to model the differentiation processes of stem cells and better control them,” said Ben-Yakar.
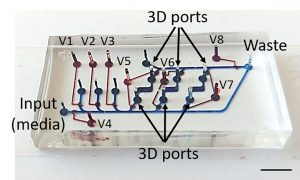
To understand how people develop from embryos and what can go wrong at those early stages, developmental biologists study the mother of all cells — stem cells. By differentiation, stem cells give rise to all the different types of cells in our bodies, from the cells in our responsive retinas to those in tough toenails.
But stem cells are a diverse bunch, dynamically turning genes on and off in a pattern that can be unique from their neighbors. To really understand cause and effect — why this or that stimulus results in a specific cellular response — each stem cell requires separate examination. High-resolution microscopy is perfect for individually tracking cells and their cellular players, but it is technically challenging to perform long-term imaging while manipulating the temperamental stem cells.
Existing two-layer microfluidic devices have control valves in the lower layer that restrict cell culture to the upper layer. This introduces a gap between cells and the thin glass substrate, which makes high-resolution microscopy impossible. But the team designed new 3D ports with upper layer valves controlling fluid flow between layers. This enabled cells to be cultured directly on the glass of the lower layer for optimal imaging.
Ben-Yakar explained it was a long-iterative process to find the optimum conditions for cell growth within such a low-volume, specialized environment. Eventually, they settled on the timing for nutrient broth exchange, which was not too regularly. Otherwise, cells can’t signal to one another and don’t grow.
They also looked at how fast fluid exchange could occur without stressing the cells. Once the team were able to reliably and repeatedly culture healthy stem cells, they demonstrated the high-resolution imaging of fluorescent markers of stem cell stability.
“For me, it’s very important to develop a user friendly platform that can be easily deployed in biology laboratories,” said Ben-Yakar, who started up a company — Newormics — in 2016, to advance the robustness and reliability of microfluidic devices. Next on Ben-Yakar’s list is to simplify the valve-control software of the new device and get it ready to send out to collaborators and to be used to better understand differentiation.
###
For more information:
Larry Frum
media@aip.org
301-209-3090
Article Title
An automated microfluidic device for time-lapse imaging of mouse embryonic stem cells
Authors
Adam F. Laing, Venkat Tirumala, Evan Hegarty, Sudip Mondal, Peisen Zhao, William Hamilton, Joshua M. Brickman and Adela Ben-Yakar
Author Affiliations
The University of Texas at Austin, University of Copenhagen
